
 |
Metadata Standards and RDF |
Metadata are the data which describe resources of interest. For instance, metadata about an image describe the circumstances under which the image was captured and the intellectual property details associated with the image. Metadata about an organism describe its taxonomy and details of its occurrences. An image with rich metadata has value beyond its visual characteristics because its context is known. Full metadata are available in human-readible form for each image and documented organism on the web page that is associated with it and which is returned when its HTTP URI is dereferenced in a web browser. However, full metada are also available in machine-readible form as Resource Description Framework (RDF) serialized as XML.
RDF is used to describe properties of resources (entities of interest, such as images and organisms) and the
relationships between them. For example, the following RDF fragment (in Turtle
syntax) describes the tree having the identifier http://bioimages.vanderbilt.edu/vanderbilt/7-314 using four RDF statements
(called triples):
<http://bioimages.vanderbilt.edu/vanderbilt/7-314>
rdf:type dcterms:PhysicalResource;
dwc:establishmentMeans "native"@en;
dwcuri:inCollection <http://biocol.org/urn:lsid:biocol.org:col:35259>;
foaf:depiction <http://bioimages.vanderbilt.edu/baskauf/79649>.
In machine-readible form, these triples state the kind of thing the tree is (a physical resource), how it got there
(native establishment), the collection it is part of (the Vanderbilt Arboretum),
and that it is depicted by a particular image. These last two statements link the tree's metadata to RDF descriptions
of the related resources (the arboretum and the image) in accordance with Linked Data principles.
The web page for each image and individual organism contains a link to the RDF formatted metadata for that particular
resource (in XML syntax).
It does little good to provide machine-readible metadata about a resource if the terms used to specify the properties of the resource are not standardized. Standard metadata terms provide a consistent language for expressing metadata as RDF. The Darwin Core (DwC) and Audiovisual Core (AC) TDWG standards combined with terms from the Dublin Core vocabulary provide many of the metadata terms necessary to describe live organism images and the plants that they document. Wherever possible, Bioimages uses terms from these vocabularies to describe resources in the collection.
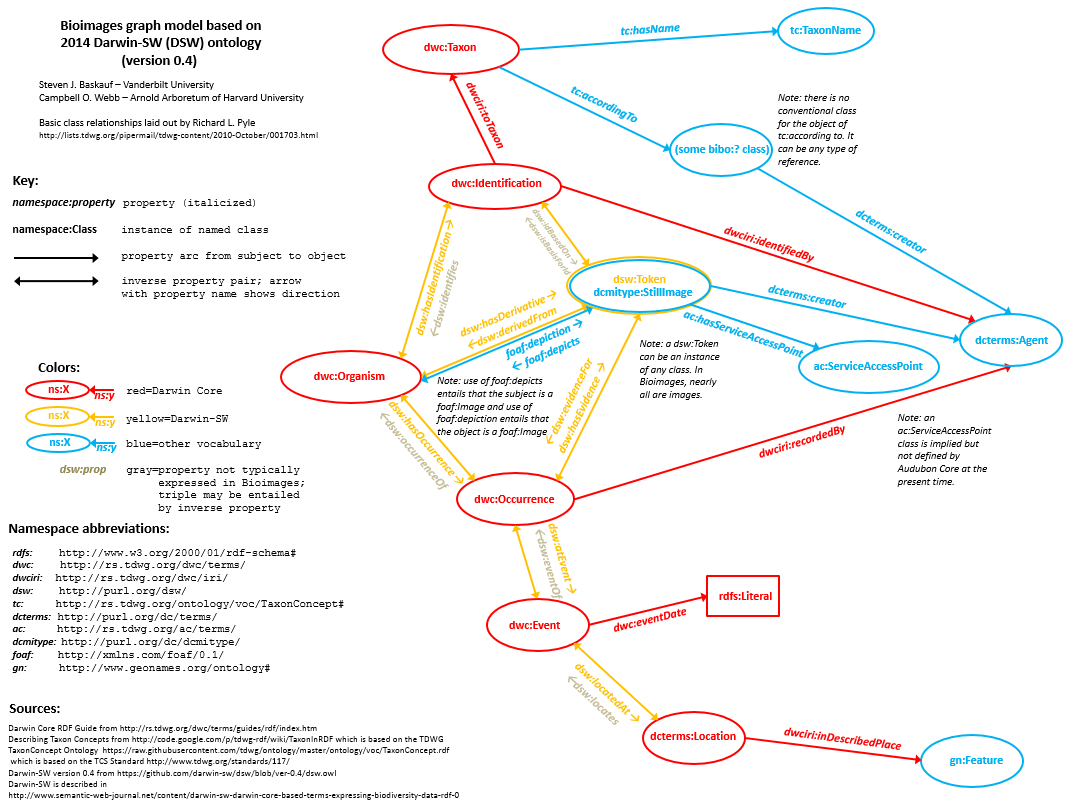
The Darwin Core TDWG standard contains many terms to describe data properties of biodiversity resources. It does not generally provide terms to describe the relationships among different types of resources. The Darwin-SW (DSW=Darwin Semantic Web) ontology expands upon the basic DwC vocabulary by formally defining the relationships among resource classes. Bioimages uses DSW object properties to describe RDF relationships between resources. A significant feature of DSW is that a living organism acts as a node which connects all occurrences derived from the individual as well as one or more taxonomic determinations as described in Baskauf (2010), an approach that is now supported by the addition of the Darwin Core organism class (dwc:Organism). The Darwin-SW model permits the expression of complex relationships in RDF that would be difficult to model in a simple database table and the Bioimages graph model is based primarily on the Darwin-SW model.
A semantic client ("machine"; computer software) can acquire Bioimages RDF/XML by dereferencing particular
image and organism HTTP URIs. The client may discover a URI via a link external to Bioimages or through
the individual organism or image RDF site maps which are
linked to the site's VOID description. The HTML content is also indexed in a
site map file which links to a subset of the RDF in the form of RDFa encoded
in the HTML of the static web pages.
The entire Bioimages database as RDF (over one million
triples) is available via the Bioimages GitHub repository in RDF/XML format as a compressed file (bioimages-rdf.zip).
These triples can also be queried via the Vanderbilt Heard Libraries SPARQL endpoint at https://sparql.vanderbilt.edu/sparql.
Describing SPARQL is beyond the scope of this web page, but see the references below for more information.